Dr. Shan ZHANG 張珊
Biography
Dr. Shan Zhang is a microbiologist and bioinformatician. She obtained her Ph.D. in Microbiology and Immunology from the School of Biotechnology and Biomolecular Sciences at the University of New South Wales (UNSW), Sydney, Australia, in 2022. During her Ph.D. program, she visited the Medical Systems Biology Lab at Christian-Albrechts-University in Kiel, Germany, in 2019 as a visiting scholar. After completing her Ph.D. degree, she pursued postdoctoral fellowships at UNSW (from March 2022 to July 2023) and at HKUST (from August 2024 to March 2025). In March 2025, she joined the Department of Pharmacology and Pharmacy, LKS Faculty of Medicine at the University of Hong Kong (HKU), as a Research Assistant Professor.
During her Ph.D. program and postdoctoral research, her projects focused on the evolutionary adaptation of microbial symbionts to a symbiotic lifestyle, particularly through the integration of multi-omics approaches (e.g., metagenomics and metabolomics) to study sponges, the common ancestor of all animals, which allows to identify ancestral and conserved features in animal host-microbiome symbioses. Her expertise in cutting-edge bioinformatics techniques, such as genome-scale metabolic modeling, facilitates the exploration of metabolic interactions within host-microbiome symbioses. Additionally, she excels in integrating both culture-independent and culture-dependent approaches to investigate these topics and applies them to various host systems, ranging from animals (e.g., sponges and corals) to plants (e.g., seaweeds and wheat).
After joining the Department of Pharmacology and Pharmacy, Dr. Zhang’s research has focused on the symbiotic relationships between the host gut and its associated eukaryotic and prokaryotic microorganisms, aiming to better understand host health and microbial physiology. She is currently involved in two research projects:
- Discovery of novel drugs from natural bioactive products. This project integrates metabolomics and machine learning to identify both known and novel bioactive compounds from marine sponges—one of the richest sources of new marine natural products reported annually, offering a diverse array of biologically significant compounds.
- Dysregulated gut-microbiome co-metabolism in metabolic-associated fatty liver disease (MAFLD). This project employs metabolic modeling approaches to investigate the interactions between the host gut and the microbiome in the context of MAFLD. Ultimately, machine learning will be applied to provide insights in disease prediction based on the state-specific microbial signatures.
The research projects will be approached from the following perspectives:
- Identifying key players in the microbial community that underpins host fitness.
- Uncovering the essential roles of microbiome in nutrient cycling.
- Isolating yet-uncultured beneficial microorganisms associated with host health.
- Investigating colonization mechanisms of microorganisms compared to their bulk community counterparts.
- Developing tailored growth media to enhance the production of target bioactive compounds.
- Applying machine learning to predict microbiome fingerprint features associated with metabolic-associated diseases.
Dr. Zhang has published six first-author papers, with her research featured in journals such as Nature Communications, mSystems and Microbiome and Extremophiles. Additionally, she has co-authored four publications in Nature Microbiology, Microbiome, Environmental Microbiology and Bioinformatics.
Memberships & Editorships
Memberships:- Mental Health First Aider in Hong Kong (June 2024 - now)
Honours and Awards
- Best Poster Presentation Award for Early-Career Scientists, The 17th Deep-Sea Bio Symposium (DSBS), January 2025.
- Outstanding Scientist in the 2024 Western Pacific Cruise, 2021-2030 United Nations Decade of Ocean Science for Sustainable Development Project, August 2024.
- Ph.D. Top-Up Scholarship, UNSW, Australia, 2017-2019.
- Travel awards, The 18th Conference of International Society for Microbial Ecology (ISME18), May 2022
- Higher Degree Research Completion Scholarship, UNSW, Australia, 2021
- Science Ph.D. Writing Scholarship, UNSW, Australia, 2020
- Adrian Lee Travel Scholarship, UNSW, Australia, 2019
Research Interests
- Host-microbe symbioses
- Systems biology
- Metabolic modelling
- Multi-omics analysis
- Microbial physiology
- Microscopy
- Bioinformatics

Figure 1. Dr. Shan Zhang with the manned submersible JIAOLONG in the Western Pacific Ocean.
Dr. Zhang participated in the international cruise of the Digital Deep-Sea Typical Habitats Programme and dives with JIAOLONG to a depth of 1,263 meters in the Westen Pacific Ocean to investigate microbial diversity. She made history as the first female passenger in an all-female submersible dive globally. Her achievement has been featured in various media outlets, including Xinhua News, TVB News, Wen Wei Po, and Ming Pao Daily News.
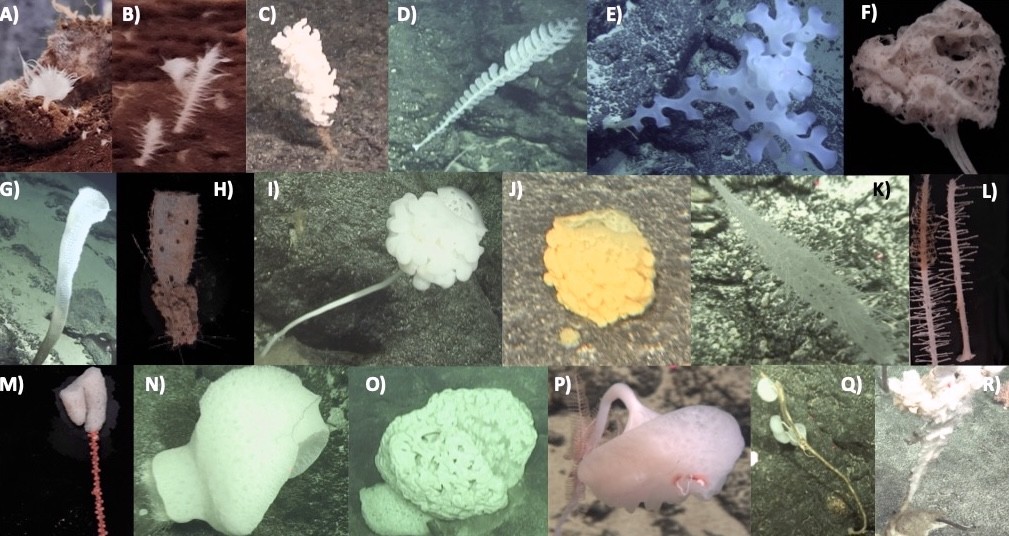
Figure 2. Gallery of deep-sea sponges collected by Dr. Shan Zhang during the international cruise of the Digital Deep-Sea Typical Habitats Programme.
These specimens serve as valuable resources for exploring marine-derived bioactive compounds with potential applications in novel drug discovery.
Publications
Publication Highlights
- Zhang, S., Song, W., Marinos, G., Waschina, S., Zimmermann, J., Kaleta, C. & Thomas, T. (2024). Genome-scale metabolic modelling reveals interactions and key roles of symbiont clades in a sponge holobiont. Nature Communication 15, 10858 (IF:14.7) https://doi.org/10.1038/s41467-024-55222-w (This study utilized a constraint-based metabolic network to predict metabolic interactions in the sponge microbiome. By integrating community structure with multi-omics data, we predicted microbial behavior under various environmental stresses using the cutting-edge genome-scale metabolic modelling approach. Notably, we identified the sponge Stylissa sp. microbiome as a carbon sink, highlighting its crucial role in global biogeochemical cycling.)
- Zhang, S., Song, W., Nothias, L. F., Couvillion, S. P., Webster, N., & Thomas, T. (2022). Comparative metabolomic analysis reveals shared and unique chemical interactions in sponge holobionts. Microbiome, 10(1), 1-14. (IF: 16.837) https://doi.org/10.1186/s40168-021-01220-9 (This study employed comparative metabolomics to identify the true producers and ecological roles of sponge-derived metabolites, which could potentially mediate metabolic integrations within the sponge holobionts.)
- Zhang, S., Song, W., Wemheuer, B., Reveillaud, J., Webster, N., & Thomas, T. (2019). Comparative genomics reveals ecological and evolutionary insights into sponge-associated Thaumarchaeota. Msystems, 4(4), e00288-19. (IF: 6.53) https://doi.org/10.1128/msystems.00288-19 (This study used comparative genomics to uncover evolutionary adaptations of sponge-associated microorganisms.)
Selected Publications
- Zhang, S., Song, W., Marinos, G., Waschina, S., Zimmermann, J., Kaleta, C. & Thomas, T. (2024). Genome-scale metabolic modelling reveals interactions and key roles of symbiont clades in a sponge holobiont. Nature Communication 15, 10858 (IF:14.7) https://doi.org/10.1038/s41467-024-55222-w
- Zhang, S., Song, W., Nothias, L. F., Couvillion, S. P., Webster, N., & Thomas, T. (2022). Comparative metabolomic analysis reveals shared and unique chemical interactions in sponge holobionts. Microbiome, 10(1), 1-14. (IF: 16.837) https://doi.org/10.1186/s40168-021-01220-9
- Zhang, S., Song, W., Wemheuer, B., Reveillaud, J., Webster, N., & Thomas, T. (2019). Comparative genomics reveals ecological and evolutionary insights into sponge-associated Thaumarchaeota. Msystems, 4(4), e00288-19. (IF: 6.53) https://doi.org/10.1128/msystems.00288-19
- Zhang, S., Song, W., Yu, M., & Lin, X. (2017). Comparative genomics analysis of five Psychrobacter strains isolated from world-wide habitats reveal high intra-genus variations. Extremophiles, 21(3), 581-589. (IF: 3.06) https://doi.org/10.1007/s00792-017-0927-1
- Shaffer, J. P., Nothias, L.-F., Thompson, L. R., Sanders, J. G., Salido, R. A., Couvillion, S. P., Brejnrod, A. D., Lejzerowicz, F., Haiminen, N., Huang, S., Lutz, H. L., Zhu, Q., Martino, C., Morton, J. T., Karthikeyan, S., Nothias-Esposito, M., Dührkop, K., Böcker, S., Kim, H. W., Aksenov, A. A., Bittremieux, W., Minich, J. J., Marotz, C., Bryant, M. M., Sanders, K., Schwartz, T., Humphrey, G., Vásquez-Baeza, Y., Tripathi, A., Parida, L., Carrieri, A. P., Beck, K. L., Das, P., González, A., McDonald, D., Ladau, J., Karst, S. M., Albertsen, M., Ackermann, G., DeReus, J., Thomas, T., Petras, D., Shade, A., Stegen, J., Song, S. J., Metz, T. O., Swafford, A. D., Dorrestein, P. C., Jansson, J. K., Gilbert, J. A., Angenant, L. T., Berry, A. M., Bittleston, L. S., Bowen, J. L., Chavarría, M., Cowan, D. A., Distel, D., Girguis, P. R., Huerta-Cepas, J., Jensen, P. R., Jiang, L., King, G. M., Lavrinienko, A., MacRae-Crerar, A., Makhalanyane, T. P., Mappes, T., Marzinelli, E. M., Mayer, G., McMahon, K. D., Metcalf, J. L., Miyake, S., Mousseau, T. A., Murillo-Cruz, C., Myrold, D., Palenik, B., Pinto-Tomás, A. A., Porazinska, D. L., Ramond, J.-B., Rowher, F., RoyChowdhury, T., Sandin, S. A., Schmidt, S. K., Seedorf, H., Shade, A., Shipway, J. R., Smith, J. E., Stegen, J., Stewart, F. J., Tait, K., Thomas, T., Tucker, Y., U’Ren, J. M., Watts, P. C., Webster, N. S., Zaneveld, J. R., Zhang, S. (2022). Standardized multi-omics of Earth’s microbiomes reveals microbial and metabolite diversity. Nature microbiology, 1-23. (IF: 30.964) https://doi.org/10.1038/s41564-022-01266-x
- Song, W., Zhang, S., & Thomas, T. (2022). MarkerMAG: linking metagenome-assembled genomes (MAGs) with 16S rRNA marker genes using paired-end short reads. Bioinformatics. (IF: 6.937) https://doi.org/10.1093/bioinformatics/btac398
- Song, W., Wemheuer, B., Zhang, S., Steensen, K., & Thomas, T. (2019). MetaCHIP: community-level horizontal gene transfer identification through the combination of best-match and phylogenetic approaches. Microbiome, 7(1), 1-14. Microbiome. 2019 Dec;7(1):36. (IF: 16.837) https://doi.org/10.1186/s40168-019-0649-y





